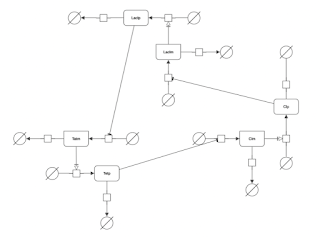
Week 13 Update - Wrapping Up
This is the last week of Google Summer of Code. I am happy to share that we have met most of the goals outlined in our proposal. I am thankful to my mentor Dr. Matthias König for his guidance and supervision throughout the summer. I hope the NRNB community finds our project useful and I'll continue to be a part of the open-source world forever. An update on the current project status: GSoC final submission is due before September 12 (Monday). Work submission guidelines are here . We must move the complete code from the libsbml-notebook branch to the main branch. Github CI is failing. We need to sort it out. Combine archive created. Need review. LibOmexMeta - too complex; I think we can skip this. Hosting notebooks on google colab. Need review. Pending code tasks: CellML: Code written, some minor issues to discuss: math equations, the syntax for public_interface, and connections between two components. SBOL: Waiting for response from Gonzalo. Standard Co...